Chapter 8 Data
Has it occurred to you that she might not have been a reliable source of information?
— Jon Snow
With the knowledge acquired in previous chapters, you are now equipped to start doing analysis and modeling at scale! So far, however, we haven’t really explained much about how to read data into Spark. We’ve explored how to use copy_to() to upload small datasets or functions like spark_read_csv() or spark_write_csv() without explaining in detail how and why.
So, you are about to learn how to read and write data using Spark. And, while this is important on its own, this chapter will also introduce you to the data lake—a repository of data stored in its natural or raw format that provides various benefits over existing storage architectures. For instance, you can easily integrate data from external systems without transforming it into a common format and without assuming those sources are as reliable as your internal data sources.
In addition, we will also discuss how to extend Spark’s capabilities to work with data not accessible out of the box and make several recommendations focused on improving performance for reading and writing data. Reading large datasets often requires you to fine-tune your Spark cluster configuration, but that’s the topic of Chapter 9.
8.1 Overview
In Chapter 1, you learned that beyond big data and big compute, you can also use Spark to improve velocity, variety, and veracity in data tasks. While you can use the learnings of this chapter for any task requiring loading and storing data, it is particularly interesting to present this chapter in the context of dealing with a variety of data sources. To understand why, we should first take a quick detour to examine how data is currently processed in many organizations.
For several years, it’s been a common practice to store large datasets in a relational database, a system first proposed in 1970 by Edgar F. Codd.21 You can think of a database as a collection of tables that are related to one another, where each table is carefully designed to hold specific data types and relationships to other tables. Most relational database systems use Structured Query Language (SQL) for querying and maintaining the database. Databases are still widely used today, with good reason: they store data reliably and consistently; in fact, your bank probably stores account balances in a database and that’s a good practice.
However, databases have also been used to store information from other applications and systems. For instance, your bank might also store data produced by other banks, such as incoming checks. To accomplish this, the external data needs to be extracted from the external system, transformed into something that fits the current database, and finally be loaded into it. This is known as Extract, Transform, and Load (ETL), a general procedure for copying data from one or more sources into a destination system that represents the data differently from the source. The ETL process became popular in the 1970s.
Aside from databases, data is often also loaded into a data warehouse, a system used for reporting and data analysis. The data is usually stored and indexed in a format that increases data analysis speed but that is often not suitable for modeling or running custom distributed code. The challenge is that changing databases and data warehouses is usually a long and delicate process, since data needs to be reindexed and the data from multiple data sources needs to be carefully transformed into single tables that are shared across data sources.
Instead of trying to transform all data sources into a common format, you can embrace this variety of data sources in a data lake, a system or repository of data stored in its natural format (see Figure 8.1). Since data lakes make data available in its original format, there is no need to carefully transform it in advance; anyone can use it for analysis, which adds significant flexibility over ETL. You then can use Spark to unify data processing from data lakes, databases, and data warehouses through a single interface that is scalable across all of them. Some organizations also use Spark to replace their existing ETL process; however, this falls in the realm of data engineering, which is well beyond the scope of this book. We illustrate this with dotted lines in Figure 8.1.
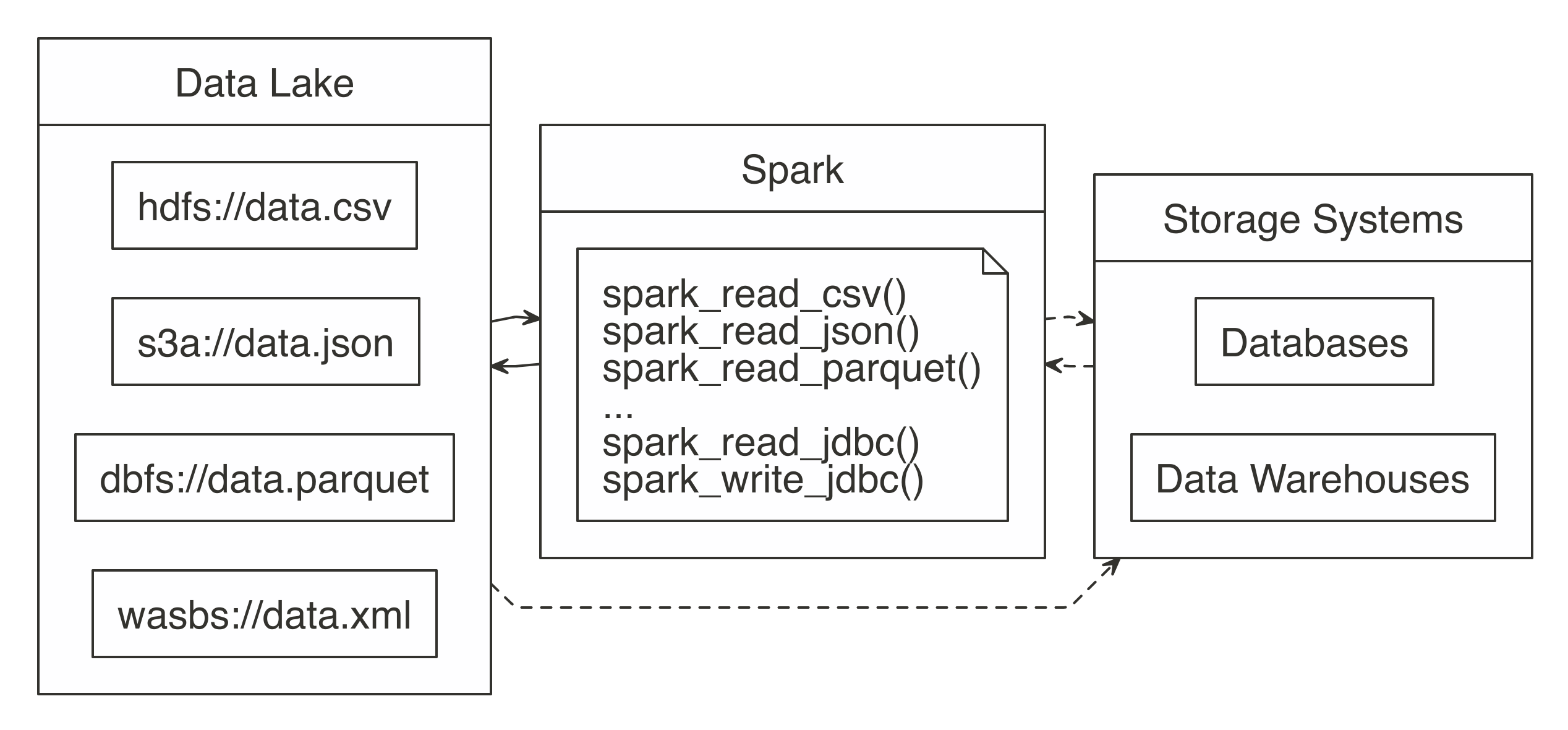
FIGURE 8.1: Spark processing raw data from a data lakes, databases, and data warehouses
In order to support a broad variety of data source, Spark needs to be able to read and write data in several different file formats (CSV, JSON, Parquet, etc), access them while stored in several file systems (HDFS, S3, DBFS, etc) and, potentially, interoperate with other storage systems (databases, data warehouses, etc). We will get to all of that; but first, we will start by presenting how to read, write and copy data using Spark.
8.2 Reading Data
To support a broad variety of data sources, Spark needs to be able to read and write data in several different file formats (CSV, JSON, Parquet, and others), and access them while stored in several file systems (HDFS, S3, DBFS, and more) and, potentially, interoperate with other storage systems (databases, data warehouses, etc.). We will get to all of that, but first, we will start by presenting how to read, write, and copy data using Spark.
8.2.1 Paths
If you are new to Spark, it is highly recommended to review this section before you start working with large datasets. We will introduce several techniques that improve the speed and efficiency of reading data. Each subsection presents specific ways to take advantage of how Spark reads files, such as the ability to treat entire folders as datasets as well as being able to describe them to read datasets faster in Spark.
letters <- data.frame(x = letters, y = 1:length(letters))
dir.create("data-csv")
write.csv(letters[1:3, ], "data-csv/letters1.csv", row.names = FALSE)
write.csv(letters[1:3, ], "data-csv/letters2.csv", row.names = FALSE)
do.call("rbind", lapply(dir("data-csv", full.names = TRUE), read.csv)) x y
1 a 1
2 b 2
3 c 3
4 a 1
5 b 2
6 c 3In Spark, there is the notion of a folder as a dataset. Instead of enumerating each file, simply pass the path containing all the files. Spark assumes that every file in that folder is part of the same dataset. This implies that the target folder should be used only for data purposes. This is especially important since storage systems like HDFS store files across multiple machines, but, conceptually, they are stored in the same folder; when Spark reads the files from this folder, it’s actually executing distributed code to read each file within each machine—no data is transferred between machines when distributed files are read:
library(sparklyr)
sc <- spark_connect(master = "local", version = "2.3")
spark_read_csv(sc, "data-csv/")# Source: spark<datacsv> [?? x 2]
x y
<chr> <int>
1 a 1
2 b 2
3 c 3
4 d 4
5 e 5
6 a 1
7 b 2
8 c 3
9 d 4
10 e 5The “folder as a table” idea is found in other open source technologies as well. Under the hood, Hive tables work the same way. When you query a Hive table, the mapping is done over multiple files within the same folder. The folder’s name usually matches the name of the table visible to the user.
Next, we will present a technique that allows Spark to read files faster as well as to reduce read failures by describing the structure of a dataset in advance.
8.2.2 Schema
When reading data, Spark is able to determine the data source’s column names and column types, also known as the schema. However, guessing the schema comes at a cost; Spark needs to do an initial pass on the data to guess what it is. For a large dataset, this can add a significant amount of time to the data ingestion process, which can become costly even for medium-size datasets. For files that are read over and over again, the additional read time accumulates over time.
To avoid this, Spark allows you to provide a column definition by providing a columns argument to describe your dataset. You can create this schema by sampling a small portion of the original file yourself:
x y
"factor" "integer" Or, you can set the column specification to a vector containing the column types explicitly. The vector’s values are named to match the field names:
x y
"character" "numeric" The accepted variable types are: integer, character, logical, double, numeric, factor, Date, and POSIXct.
Then, when reading using spark_read_csv(), you can pass spec_with_r to the columns argument to match the names and types of the original file. This helps to improve performance since Spark will not need to determine the column types.
# Source: spark<datacsv> [?? x 2]
x y
<chr> <int>
1 a 1
2 b 2
3 c 3
4 a 1
5 b 2
6 c 3The following example shows how to set the field type to something different. However, the new field type needs to be a compatible type in the original dataset. For example, you cannot set a character field to numeric. If you use an incompatible type, the file read will fail with an error. Additionally, the following example also changes the names of the original fields:
spec_compatible <- c(my_letter = "character", my_number = "character")
spark_read_csv(sc, "data-csv/", columns = spec_compatible)# Source: spark<datacsv> [?? x 2]
my_letter my_number
<chr> <chr>
1 a 1
2 b 2
3 c 3
4 a 1
5 b 2
6 c 3 In Spark, malformed entries can cause errors during reading, particularly for non-character fields. To prevent such errors, we can use a file specification that imports them as characters and then use dplyr to coerce the field into the desired type.
This subsection reviewed how we can read files faster and with fewer failures, which lets us start our analysis more quickly. Another way to accelerate our analysis is by loading less data into Spark memory, which we examine in the next section.
8.2.3 Memory
By default, when using Spark with R, when you read data, it is copied into Spark’s distributed memory, making data analysis and other operations very fast. There are cases, such as when the data is too big, for which loading all the data might not be practical or even necessary. For those cases, Spark can just “map” the files without copying data into memory.
The mapping creates a sort of virtual table in Spark. The implication is that when a query runs against that table, Spark needs to read the data from the files at that time. Any consecutive reads after that will do the same. In effect, Spark becomes a pass-through for the data. The advantage of this method is that there is almost no up-front time cost to “reading” the file; the mapping is very fast. The downside is that running queries that actually extract data will take longer.
This is controlled by the memory argument of the read functions. Setting it to FALSE prevents the data copy (the default is TRUE):
There are good use cases for this method, one of which is when not all columns of a table are needed. For example, take a very large file that contains many columns. Assuming this is not the first time you interact with this data, you would know what columns are needed for the analysis. When you know which columns you need, the files can be read using memory = FALSE, and then the needed columns can be selected with dplyr. The resulting dplyr variable can then be cached into memory, using the compute() function. This will make Spark query the file(s), pull the selected fields, and copy only that data into memory. The result is an in-memory table that took comparatively less time to ingest:
The next section covers a short technique to make it easier to carry the original field names of imported data.
8.2.4 Columns
Spark 1.6 required that column names be sanitized, so R does that by default. There might be cases when you would like to keep the original names intact, or when working with Spark version 2.0 or above. To do that, set the sparklyr.sanitize.column.names option to FALSE:
# Source: table<iris> [?? x 5]
# Database: spark_connection
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <chr>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# ... with more rowsWith this review of how to read data into Spark, we move on to look at how we can write data from our Spark session.
8.3 Writing Data
Some projects require that new data generated in Spark be written back to a remote source. For example, the data could be new predicted values returned by a Spark model. The job processes the mass generation of predictions, but then the predictions need to be stored. This section focuses on how you should use Spark for moving the data from Spark into an external destination.
Many new users start by downloading Spark data into R, and then upload it to a target, as illustrated in Figure 8.2. It works for smaller datasets, but it becomes inefficient for larger ones. The data typically grows in size to the point that it is no longer feasible for R to be the middle point.
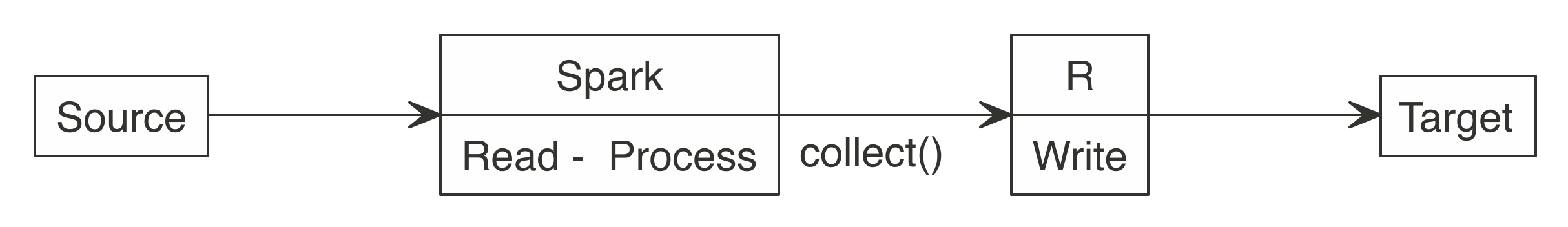
FIGURE 8.2: Incorrect use of Spark when writing large datasets
All efforts should be made to have Spark connect to the target location. This way, reading, processing, and writing happens within the same Spark session.
As Figure 8.3 shows, a better approach is to use Spark to read, process, and write to the target. This approach is able to scale as big as the Spark cluster allows, and prevents R from becoming a choke point.
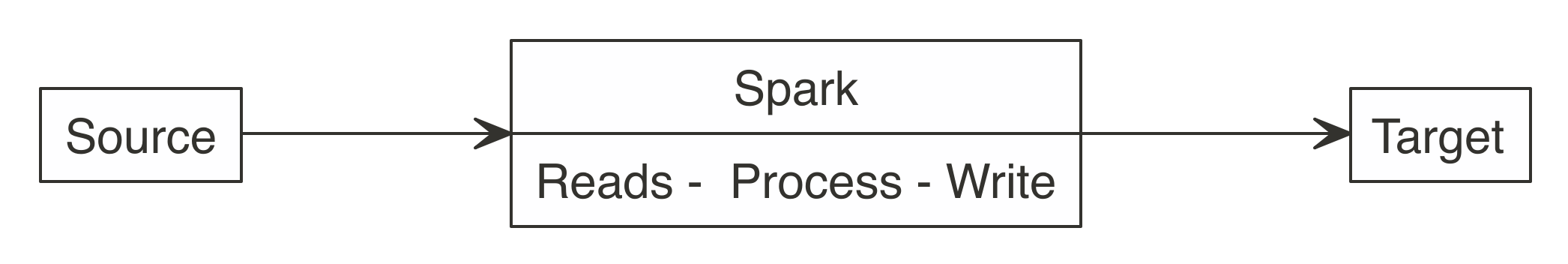
FIGURE 8.3: Correct use of Spark when writing large datasets
Consider the following scenario: a Spark job just processed predictions for a large dataset, resulting in a considerable amount of predictions. Choosing a method to write results will depend on the technology infrastructure you are working on. More specifically, it will depend on Spark and the target running, or not, in the same cluster.
Back to our scenario, we have a large dataset in Spark that needs to be saved. When Spark and the target are in the same cluster, copying the results is not a problem; the data transfer is between RAM and disk of the same cluster or efficiently shuffled through a high-bandwidth connection.
But what to do if the target is not within the Spark cluster? There are two options, and choosing one will depend on the size of the data and network speed:
- Spark transfer
- In this case, Spark connects to the remote target location and copies the new data. If this is done within the same datacenter, or cloud provider, the data transfer could be fast enough to have Spark write the data directly.
- External transfer and otherwise
- Spark can write the results to disk and transfers them via a third-party application. Spark writes the results as files and then a separate job copies the files over. In the target location, you would use a separate process to transfer the data into the target location.
It is best to recognize that Spark, R, and any other technology are tools. No tool can do everything, nor should be expected to. Next we will describe how to copy data into Spark or collect large datasets that don’t fit in-memory, this can be used to transfer data across clusters, or help initialize your distributed datasets.
8.4 Copy
Previous chapters used copy_to() as a handy helper to copy data into Spark; however, you can use copy_to() only to transfer in-memory datasets that are already loaded in memory. These datasets tend to be much smaller than the kind of datasets you would want to copy into Spark.
For instance, suppose that we have a 3 GB dataset generated as follows:
dir.create("largefile.txt")
write.table(matrix(rnorm(10 * 10^6), ncol = 10), "largefile.txt/1",
append = T, col.names = F, row.names = F)
for (i in 2:30)
file.copy("largefile.txt/1", paste("largefile.txt/", i))If we had only 2 GB of memory in the driver node, we would not be able to load this 3 GB file into memory using copy_to(). Instead, when using the HDFS as storage in your cluster, you can use the hadoop command-line tool to copy files from disk into Spark from the terminal as follows. Notice that the following code works only in clusters using HDFS, not in local environments.
You then can read the uploaded file, as described in the File Formats section; for text files, you would run:
# Source: spark<largefile> [?? x 1]
line
<chr>
1 0.0982531064914565 -0.577567317599452 -1.66433938237253 -0.20095089489…
2 -1.08322304504007 1.05962389624635 1.1852771207729 -0.230934710049462 …
3 -0.398079835552421 0.293643382374479 0.727994248743204 -1.571547990532…
4 0.418899768227183 0.534037617828835 0.921680317620166 -1.6623094393911…
5 -0.204409401553028 -0.0376212693728992 -1.13012269711811 0.56149527218…
6 1.41192628218417 -0.580413572014808 0.727722566256326 0.5746066486689 …
7 -0.313975036262443 -0.0166426329807508 -0.188906975208319 -0.986203251…
8 -0.571574679637623 0.513472254005066 0.139050812059352 -0.822738334753…
9 1.39983023148955 -1.08723592838627 1.02517804413913 -0.412680186313667…
10 0.6318328148434 -1.08741784644221 -0.550575696474202 0.971967251067794…
# … with more rowscollect() has a similar limitation in that it can collect only datasets that fit your driver memory; however, if you had to extract a large dataset from Spark through the driver node, you could use specialized tools provided by the distributed storage. For HDFS, you would run the following:
Alternatively, you can also collect datasets that don’t fit in memory by providing a callback to collect(). A callback is just an R function that will be called over each Spark partition. You then can write this dataset to disk or push to other clusters over the network.
You could use the following code to collect 3 GB even if the driver node collecting this dataset had less than 3 GB of memory. That said, as Chapter 3 explains, you should avoid collecting large datasets into a single machine since this creates a significant performance bottleneck. For conciseness, we will collect only the first million rows; feel free to remove head(10^6) if you have a few minutes to spare:
dir.create("large")
spark_read_text(sc, "largefile.txt", memory = FALSE) %>%
head(10^6) %>%
collect(callback = function(df, idx) {
writeLines(df$line, paste0("large/large-", idx, ".txt"))
})Make sure you clean up these large files and empty your recycle bin as well:
In most cases, data will already be stored in the cluster, so you should not need to worry about copying large datasets; instead, you can usually focus on reading and writing different file formats, which we describe next.
8.5 File Formats
Out of the box, Spark is able to interact with several file formats, like CSV, JSON, LIBSVM, ORC, and Parquet. Table 8.1 maps the file format to the function you should use to read and write data in Spark.
| Format | Read | Write |
|---|---|---|
| Comma separated values (CSV) | spark_read_csv() | spark_write_csv() |
| JavaScript Object Notation (JSON) | spark_read_json() | spark_write_json() |
| Library for Support Vector Machines (LIBSVM) | spark_read_libsvm() | spark_write_libsvm() |
| Optimized Row Columnar (ORC) | spark_read_orc() | spark_write_orc() |
| Apache Parquet | spark_read_parquet() | spark_write_parquet() |
| Text | spark_read_text() | spark_write_text() |
The following sections will describe special considerations particular to each file format as well as some of the strengths and weaknesses of some popular file formats, starting with the well-known CSV file format.
8.5.1 CSV
The CSV format might be the most common file type in use today. It is defined by a text file separated by a given character, usually a comma. It should be pretty straightforward to read CSV files; however, it’s worth mentioning a couple techniques that can help you process CSVs that are not fully compliant with a well-formed CSV file. Spark offers the following modes for addressing parsing issues:
- Permissive
- Inserts NULL values for missing tokens
- Drop Malformed
- Drops lines that are malformed
- Fail Fast
- Aborts if it encounters any malformed line
You can use these in sparklyr by passing them inside the options argument. The following example creates a file with a broken entry. It then shows how it can be read into Spark:
## Creates bad test file
writeLines(c("bad", 1, 2, 3, "broken"), "bad.csv")
spark_read_csv(
sc,
"bad3",
"bad.csv",
columns = list(foo = "integer"),
options = list(mode = "DROPMALFORMED"))# Source: spark<bad3> [?? x 1]
foo
<int>
1 1
2 2
3 3Spark provides an issue tracking column, which was hidden by default. To enable it, add _corrupt_record to the columns list. You can combine this with the use of the PERMISSIVE mode. All rows will be imported, invalid entries will receive an NA, and the issue will be tracked in the _corrupt_record column:
spark_read_csv(
sc,
"bad2",
"bad.csv",
columns = list(foo = "integer", "_corrupt_record" = "character"),
options = list(mode = "PERMISSIVE")
)# Source: spark<bad2> [?? x 2]
foo `_corrupt_record`
<int> <chr>
1 1 NA
2 2 NA
3 3 NA
4 NA broken Reading and storing data as CSVs is quite common and supported across most systems. For tabular datasets, it is still a popular option, but for datasets containing nested structures and nontabular data, JSON is usually preferred.
8.5.2 JSON
JSON is a file format originally derived from JavaScript that has grown to be language-independent and very popular due to its flexibility and ubiquitous support. Reading and writing JSON files is quite straightforward:
writeLines("{'a':1, 'b': {'f1': 2, 'f3': 3}}", "data.json")
simple_json <- spark_read_json(sc, "data.json")
simple_json# Source: spark<data> [?? x 2]
a b
<dbl> <list>
1 1 <list [2]>However, when you deal with a dataset containing nested fields like the one from this example, it is worth pointing out how to extract nested fields. One approach is to use a JSON path, which is a domain-specific syntax commonly used to extract and query JSON files. You can use a combination of get_json_object() and to_json() to specify the JSON path you are interested in. To extract f1 you would run the following transformation:
# Source: spark<?> [?? x 3]
a b z
<dbl> <list> <chr>
1 1 <list [2]> 2 Another approach is to install sparkly.nested from CRAN with install.packages("sparklyr.nested") and then unnest nested data with sdf_unnest():
# Source: spark<?> [?? x 3]
a f1 f3
<dbl> <dbl> <dbl>
1 1 2 3While JSON and CSVs are quite simple to use and versatile, they are not optimized for performance; however, other formats like ORC, AVRO, and Parquet are.
8.5.3 Parquet
Apache Parquet, Apache ORC, and Apache AVRO are all file formats designed with performance in mind. Parquet and ORC store data in columnar format, while AVRO is row-based. All of them are binary file formats, which reduces storage space and improves performance. This comes at the cost of making them a bit more difficult to read by external systems and libraries; however, this is usually not an issue when used as intermediate data storage within Spark.
To illustrate this, Figure 8.4 plots the result of running a 1-million-row write-speed benchmark using the bench package; feel free to use your own benchmarks over meaningful datasets when deciding which format best fits your needs:
numeric <- copy_to(sc, data.frame(nums = runif(10^6)))
bench::mark(
CSV = spark_write_csv(numeric, "data.csv", mode = "overwrite"),
JSON = spark_write_json(numeric, "data.json", mode = "overwrite"),
Parquet = spark_write_parquet(numeric, "data.parquet", mode = "overwrite"),
ORC = spark_write_parquet(numeric, "data.orc", mode = "overwrite"),
iterations = 20
) %>% ggplot2::autoplot()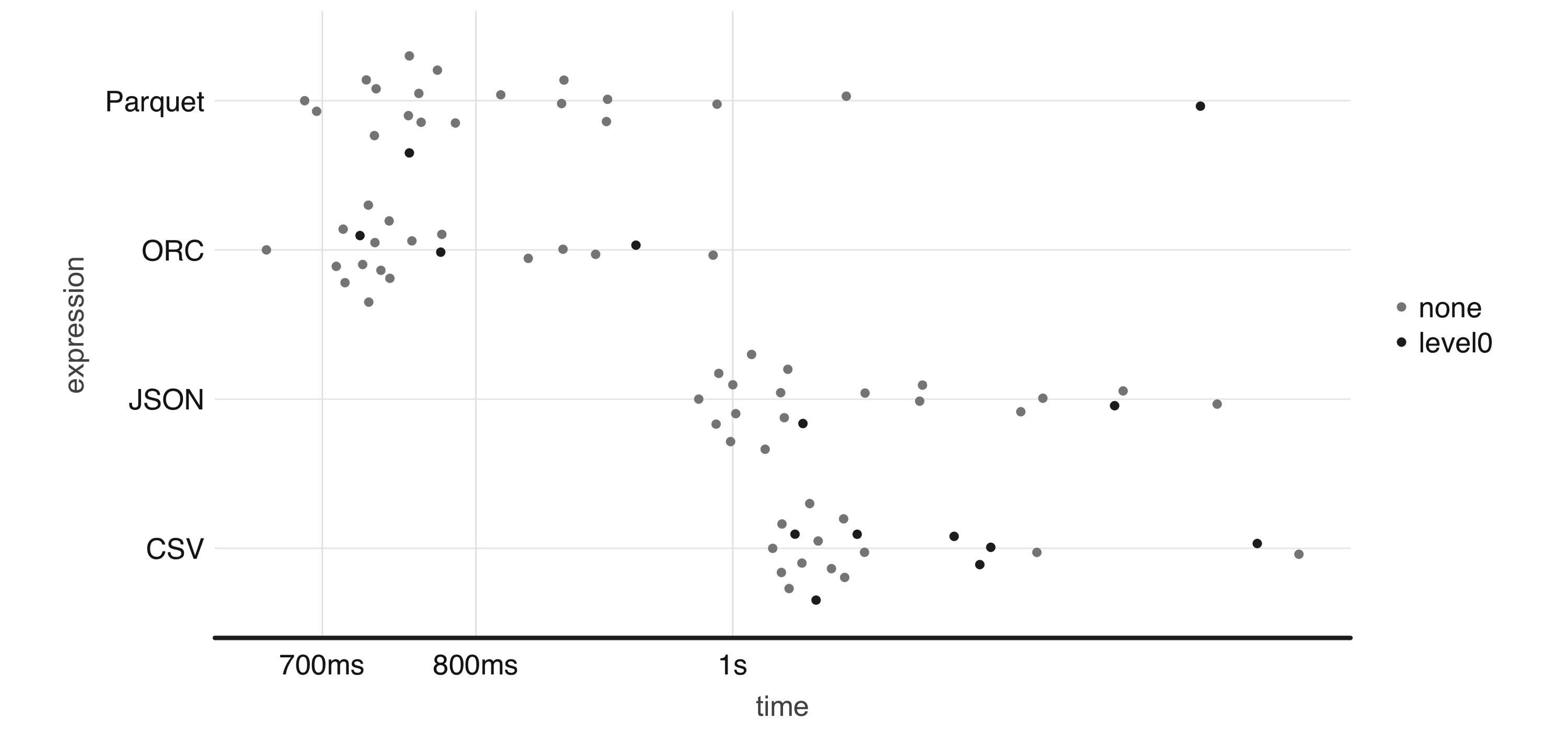
FIGURE 8.4: One-million-rows write benchmark between CSV, JSON, Parquet, and ORC
From now on, be sure to disconnect from Spark whenever we present a new spark_connect() command:
This concludes the introduction to some of the out-of-the-box supported file formats, we will present next how to deal with formats that require external packages and customization.
8.5.4 Others
Spark is a very flexible computing platform. It can add functionality by using extension programs, called packages. You can access a new source type or file system by using the appropriate package.
Packages need to be loaded into Spark at connection time. To load the package, Spark needs its location, which could be inside the cluster, in a file share, or the internet.
In sparklyr, the package location is passed to spark_connect(). All packages should be listed in the sparklyr.connect.packages entry of the connection configuration.
It is possible to access data source types that we didn’t previously list. Loading the appropriate default package for Spark is the first of two steps The second step is to actually read or write the data. The spark_read_source() and spark_write_source() functions do that. They are generic functions that can use the libraries imported by a default package.
For instance, we can read XML files as follows:
sc <- spark_connect(master = "local", version = "2.3", config = list(
sparklyr.connect.packages = "com.databricks:spark-xml_2.11:0.5.0"))
writeLines("<ROWS><ROW><text>Hello World</text></ROW>", "simple.xml")
spark_read_source(sc, "simple_xml", "simple.xml", "xml")# Source: spark<data> [?? x 1]
text
<chr>
1 Hello Worldwhich you can also write back to XML with ease, as follows:
In addition, there are a few extensions developed by the R community to load additional file formats, such as sparklyr.nested to assist with nested data, spark.sas7bdat to read data from SAS, sparkavro to read data in AVRO format, and sparkwarc to read WARC files, which use extensibility mechanisms introduced in Chapter 10. Chapter 11 presents techniques to use R packages to load additional file formats, and Chapter 13 presents techniques to use Java libraries to complement this further. But first, let’s explore how to retrieve and store files from several different file systems.
8.6 File Systems
Spark defaults to the file system on which it is currently running. In a YARN managed cluster, the default file system will be HDFS. An example path of /home/user/file.csv will be read from the cluster’s HDFS folders, not the Linux folders. The operating system’s file system will be accessed for other deployments, such as Standalone, and sparklyr’s local.
The file system protocol can be changed when reading or writing. You do this via the path argument of the sparklyr function. For example, a full path of _file://home/user/file.csv_ forces the use of the local operating system’s file system.
There are many other file system protocols, such as _dbfs://_ for Databricks’ file system, _s3a://_ for Amazon’s S3 service, _wasb://_ for Microsoft Azure storage, and _gs://_ for Google storage.
Spark does not provide support for all them directly; instead, they are configured as needed. For instance, accessing the “s3a” protocol requires adding a package to the sparklyr.connect.packages configuration setting, while connecting and specifying appropriate credentials might require using the AWS_ACCESS_KEY_ID and AWS_SECRET_ACCESS_KEY environment variables.
Sys.setenv(AWS_ACCESS_KEY_ID = my_key_id)
Sys.setenv(AWS_SECRET_ACCESS_KEY = my_secret_key)
sc <- spark_connect(master = "local", version = "2.3", config = list(
sparklyr.connect.packages = "org.apache.hadoop:hadoop-aws:2.7.7"))
my_file <- spark_read_csv(sc, "my-file", path = "s3a://my-bucket/my-file.csv")Accessing other file protocols requires loading different packages, although, in some cases, the vendor providing the Spark environment might load the package for you. Please refer to your vendor’s documentation to find out whether that is the case.
8.7 Storage Systems
A data lake and Spark usually go hand-in-hand, with optional access to storage systems like databases and data warehouses. Presenting all the different storage systems with appropriate examples would be quite time-consuming, so instead we present some of the commonly used storage systems.
As a start, Apache Hive is a data warehouse software that facilitates reading, writing, and managing large datasets residing in distributed storage using SQL. In fact, Spark has components from Hive built directly into its sources. It is very common to have installations of Spark or Hive side-by-side, so we will start by presenting Hive, followed by Cassandra, and then close by looking at JDBC connections.
8.7.1 Hive
In YARN managed clusters, Spark provides a deeper integration with Apache Hive. Hive tables are easily accessible after opening a Spark connection.
You can access a Hive table’s data using DBI by referencing the table in a SQL statement:
sc <- spark_connect(master = "local", version = "2.3")
spark_read_csv(sc, "test", "data-csv/", memory = FALSE)
DBI::dbGetQuery(sc, "SELECT * FROM test limit 10")Another way to reference a table is with dplyr using the tbl() function, which retrieves a reference to the table:
It is important to reiterate that no data is imported into R; the tbl() function only creates a reference. You then can pipe more dplyr verbs following the tbl() command:
Hive table references assume a default database source. Often, the needed table is in a different database within the metastore. To access it using SQL, prefix the database name to the table. Separate them using a period, as demonstrated here:
In dplyr, the in_schema() function can be used. The function is used inside the tbl() call:
You can also use the tbl_change_db() function to set the current session’s default database. Any subsequent call via DBI or dplyr will use the selected name as the default database:
The following examples require additional Spark packages and databases which might be difficult to follow unless you happen to have a JDBC driver or Cassandra database accessible to you.
Next, we explore a less structured storage system, often referred to as a NoSQL database.
8.7.2 Cassandra
Apache Cassandra is a free and open source, distributed, wide-column store, NoSQL database management system designed to handle large amounts of data across many commodity servers. While there are many other database systems beyond Cassandra, taking a quick look at how Cassandra can be used from Spark will give you insight into how to make use of other database and storage systems like Solr, Redshift, Delta Lake, and others.
The following example code shows how to use the datastax:spark-cassandra-connector package to read from Cassandra. The key is to use the org.apache.spark.sql.cassandra library as the source argument. It provides the mapping Spark can use to make sense of the data source. Unless you have a Cassandra database, skip executing the following statement:
sc <- spark_connect(master = "local", version = "2.3", config = list(
sparklyr.connect.packages = "datastax:spark-cassandra-connector:2.3.1-s_2.11"))
spark_read_source(
sc,
name = "emp",
source = "org.apache.spark.sql.cassandra",
options = list(keyspace = "dev", table = "emp"),
memory = FALSE)One of the most useful features of Spark when dealing with external databases and data warehouses is that Spark can push down computation to the database, a feature known as pushdown predicates. In a nutshell, pushdown predicates improve performance by asking remote databases smart questions. When you execute a query that contains the filter(age > 20) expression against a remote table referenced through spark_read_source() and not loaded in memory, rather than bringing the entire table into Spark, it will be passed to the remote database and only a subset of the remote table is retrieved.
While it is ideal to find Spark packages that support the remote storage system, there will be times when a package is not available and you need to consider vendor JDBC drivers.
8.7.3 JDBC
When a Spark package is not available to provide connectivity, you can consider a JDBC connection. JDBC is an interface for the programming language Java, which defines how a client can access a database.
It is quite easy to connect to a remote database with spark_read_jdbc(), and spark_write_jdbc(); as long as you have access to the appropriate JDBC driver, which at times is trivial and other times is quite an adventure. To keep this simple, we can briefly consider how a connection to a remote MySQL database could be accomplished.
First, you would need to download the appropriate JDBC driver from MySQL’s developer portal and specify this additional driver as a sparklyr.shell.driver-class-path connection option. Since JDBC drivers are Java-based, the code is contained within a JAR (Java ARchive) file. As soon as you’re connected to Spark with the appropriate driver, you can use the jdbc:// protocol to access particular drivers and databases. Unless you are willing to download and configure MySQL on your own, skip executing the following statement:
sc <- spark_connect(master = "local", version = "2.3", config = list(
"sparklyr.shell.driver-class-path" =
"~/Downloads/mysql-connector-java-5.1.41/mysql-connector-java-5.1.41-bin.jar"
))
spark_read_jdbc(sc, "person_jdbc", options = list(
url = "jdbc:mysql://localhost:3306/sparklyr",
user = "root", password = "<password>",
dbtable = "person"))If you are a customer of particular database vendors, making use of the vendor-provided resources is usually the best place to start looking for appropriate drivers.
8.8 Recap
This chapter expanded on how and why you should use Spark to connect and process a variety of data sources through a new data storage model known as data lakes—a storage pattern that provides more flexibility than standard ETL processes by enabling you to use raw datasets with, potentially, more information to enrich data analysis and modeling.
We also presented best practices for reading, writing, and copying data into and from Spark. We then returned to exploring the components of a data lake: file formats and file systems, with the former representing how data is stored, and the latter where the data is stored. You then learned how to tackle file formats and storage systems that require additional Spark packages, reviewed some of the performance trade-offs across file formats, and learned the concepts required to make use of storage systems (databases and warehouses) in Spark.
While reading and writing datasets should come naturally to you, you might still hit resource restrictions while reading and writing large datasets. To handle these situations, Chapter 9 shows you how Spark manages tasks and data across multiple machines, which in turn allows you to further improve the performance of your analysis and modeling tasks.
Codd EF (1970). “A relational model of data for large shared data banks.”↩